-Search query
-Search result
Showing all 40 items for (author: yao & yn)

PDB-8jgg: 
CryoEM structure of Gi-coupled MRGPRX1 with peptide agonist BAM8-22
Method: single particle / : Sun JP, Xu HE, Ynag F, Liu ZM, Guo LL, Zhang YM, Fang GX, Tie L, Zhuang YM, Xue CY

EMDB-29578: 
G4 RNA-mediated PRC2 dimer
Method: single particle / : Song J, Kasinath V
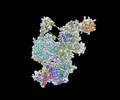
EMDB-29647: 
Body1 of G4 RNA-mediated PRC2 dimer from multibody refinement
Method: single particle / : Jiarui JS, Vignesh VK
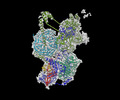
EMDB-29656: 
Body2 of G4 RNA-mediated PRC2 dimer from multibody refinement
Method: single particle / : Jiarui JS, Vignesh VK

PDB-8fyh: 
G4 RNA-mediated PRC2 dimer
Method: single particle / : Song J, Kasinath V
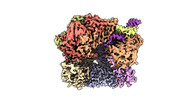
EMDB-29412: 
EM map of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 1 (open 9-1-1 and shoulder bound DNA only)
Method: single particle / : Zheng F, Georgescu R, Yao YN, O'Donnell ME, Li H
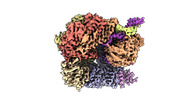
EMDB-29413: 
EM map of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 2 (open 9-1-1 ring and flexibly bound chamber DNA)
Method: single particle / : Zheng F, Georgescu R, Yao YN, O'Donnell ME, Li H
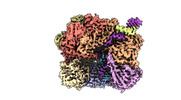
EMDB-29414: 
EM map of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 3 (open 9-1-1 and stably bound chamber DNA)
Method: single particle / : Zheng F, Georgescu R, Yao YN, O'Donnell ME, Li H
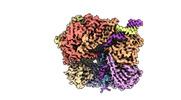
EMDB-29415: 
EM map of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 4 (partially closed 9-1-1 and stably bound chamber DNA)
Method: single particle / : Zheng F, Georgescu R, Yao YN, O'Donnell ME, Li H
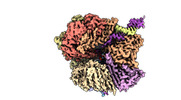
EMDB-29416: 
EM map of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 5 (closed 9-1-1 and stably bound chamber DNA)
Method: single particle / : Zheng F, Georgescu R, Yao YN, O'Donnell ME, Li H
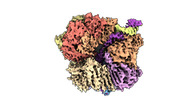
EMDB-29417: 
EM map of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 5-nt gapped DNA (9-1-1 encircling fully bound DNA)
Method: single particle / : Zheng F, Georgescu R, Yao YN, O'Donnell ME, Li H

EMDB-25872: 
Cryo-EM 3D map of the S. cerevisiae clamp-clamp loader complex PCNA-RFC bound to two DNA molecules, one at the 5'-recessed end and the other at the 3'-recessed end
Method: single particle / : Zheng F, Georgescu R, Yao YN, O'Donnell ME, Li H

EMDB-25873: 
Cryo-EM 3D map of the S. cerevisiae clamp-clamp loader complex PCNA-RFC bound to DNA with an open clamp
Method: single particle / : Zheng F, Georgescu R, Yao YN, O'Donnell ME, Li H

EMDB-25874: 
Cryo-EM 3D map of S. cerevisiae clamp-clamp loader complex PCNA-RFC bound to DNA with a closed clamp ring
Method: single particle / : Zheng F, Georgescu R, Yao YN, O'Donnell ME, Li H

EMDB-25875: 
Cryo-EM 3D map of S. cerevisiae clamp loader RFC bound to two DNA molecules, one at the 5'-recessed end and the other at the 3'-recessed end
Method: single particle / : Zheng F, Georgescu R, Yao YN, O'Donnell ME, Li H

EMDB-25876: 
Cryo-EM 3D map of S. cerevisiae clamp loader RFC bound to DNA
Method: single particle / : Zheng F, Georgescu R, Yao YN, O'Donnell ME, Li H

EMDB-26429: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the neutralizing antibody Fab fragment, C1520
Method: single particle / : Barnes CO

EMDB-26430: 
Structure of the SARS-CoV-2 NTD in complex with C1520, local refinement
Method: single particle / : Barnes CO

EMDB-26431: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the neutralizing antibody Fab fragment, C1717
Method: single particle / : Barnes CO

EMDB-26432: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the neutralizing antibody Fab fragment, C1791
Method: single particle / : Barnes CO

PDB-7uap: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the neutralizing antibody Fab fragment, C1520
Method: single particle / : Barnes CO

PDB-7uaq: 
Structure of the SARS-CoV-2 NTD in complex with C1520, local refinement
Method: single particle / : Barnes CO

PDB-7uar: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the neutralizing antibody Fab fragment, C1717
Method: single particle / : Barnes CO

EMDB-25121: 
Cryo-EM 3D map of the yeast Rad24-RFC loader bound to DNA and the closed 9-1-1 clamp
Method: single particle / : Zheng F, Georgescu R, Yao YN, O'Donnell ME, Li H

EMDB-25122: 
Cryo-EM 3D map of the yeast Rad24-RFC loader bound to DNA and the open 9-1-1 clamp
Method: single particle / : Zheng F, Georgescu R, Yao YN, O'Donnell ME, Li H

EMDB-30406: 
The LAT2-4F2hc complex in complex with tryptophan
Method: single particle / : Yan RH, Zhou JY, Li YN, Lei JL, Zhou Q

EMDB-30407: 
The LAT2-4F2hc complex in complex with leucine
Method: single particle / : Yan RH, Zhou JY, Li YN, Lei JL, Zhou Q

EMDB-21671: 
CryoEM Structure of the glucagon receptor with a dual-agonist peptide
Method: single particle / : Belousoff MJ, Sexton P, Danev R

PDB-6whc: 
CryoEM Structure of the glucagon receptor with a dual-agonist peptide
Method: single particle / : Belousoff MJ, Sexton P, Danev R

EMDB-0903: 
Heteromeric amino acid transporter b0,+AT-rBAT complex bound with Arginine
Method: single particle / : Yan RH, Li YN, Lei JL, Zhou Q

EMDB-0904: 
Heteromeric amino acid transporter b0,+AT-rBAT complex
Method: single particle / : Yan RH, Li YN, Lei JL, Zhou Q

EMDB-0905: 
cryo EM map of b0,+AT-rBAT complex, focused refined on soluble domain
Method: single particle / : Yan RH, Li YN, Lei JL, Zhou Q

EMDB-0906: 
Cryo EM map of b0,+AT-rBAT complex, focused refined on TM domain
Method: single particle / : Yan RH, Li YN, Lei JL, Zhou Q

EMDB-0907: 
Cryo EM map of b0,+AT-rBAT complex bound with Arginine, focused refined on soluble domain
Method: single particle / : Yan RH, Li YN, Lei JL, Zhou Q

EMDB-0908: 
Cryo EM map of b0,+ AT-rBAT complex bound with Arginine, focused refined on TM domain
Method: single particle / : Yan RH, Li YN, Lei JL, Zhou Q

EMDB-20074: 
Chimpanzee SIV Env trimeric ectodomain.
Method: single particle / : Pallesen J, Andrabi R, de Val N, Burton DR, Ward AB

PDB-6ohy: 
Chimpanzee SIV Env trimeric ectodomain.
Method: single particle / : Pallesen J, Andrabi R, de Val N, Burton DR, Ward AB

PDB-3j9b: 
Electron cryo-microscopy of an RNA polymerase
Method: single particle / : Chang SH, Sun DP, Liang HH, Wang J, Li J, Guo L, Wang XL, Guan CC, Boruah BM, Yuan LM, Feng F, Yang MR, Wojdyla J, Wang JW, Wang MT, Wang HW, Liu YF
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
